Here, you can find downloadable materials (or relevant links) from our research.
2021 April
AutoCoEv – a high-throughput in silico pipeline for predicting novel protein-protein interactions
AutoCoEv is a user-friendly computational pipeline for the search of co-evolution between hundreds and even thousands of proteins. Driving over 10 individual programs, with CAPS2 as a key software for co-evolution detection, AutoCoEv achieves seamless automation and parallelization of the workflow.
Project link at GitHub: https://github.com/mattilalab/autocoev/
Script authors: Petar Petrov (bash), Luqman Awoniyi (R)
2019 July
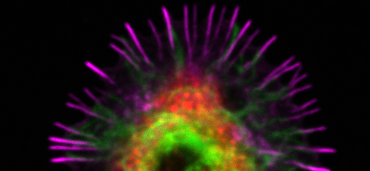
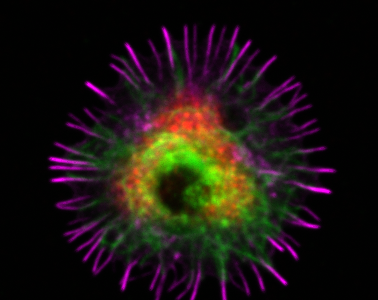
Analysis of intracellular vesicles in B lymphocytes: Antigen traffic in the spotlight
The Script should be run with FIJI (ImageJ). Upon B cell activation, antigen is rapidly internalised followed by trafficking of the antigen-containing vesicles towards a perinuclear compartment close to the MTOC. Here, we provide an example of how to use the open-source software Fiji and its 3D object counter function to quantitatively analyse the number of antigen vesicles and their distance from MTOC, as a proxy of vesicle clustering. MTOC is marked with anti-PCM1 antibodies and identified as the brightest spot in the channel.
Project link at GitHub: https://github.com/mattilalab/MiMB/tree/master/Hernandez-Perez_et_al_2019
Script author: Vid Šuštar
2019 March
Bash scripts
Scripts and other material for our recent paper titled: “Computational analysis of the evolutionarily conserved Missing In Metastasis / Metastasis Suppressor 1 gene predicts novel interactions, regulatory regions and transcriptional control” are available at GitHub.
Project link at GitHub: https://github.com/mattilalab/petrov-et-al-2019
Scripts author: Petar Petrov
For more information:
Petrov P, Sarapulov AV, Eory L, Scielzo C, Scarfo L, Smith J, Burt DW and Mattila PK. (2019) Computational analysis of the evolutionarily conserved Missing In Metastasis/Metastasis Suppressor 1 gene predicts novel interactions, regulatory regions and transcriptional control Scientific Reports. 9:4155 | https://rdcu.be/bqnHe
PubMed link: https://www.ncbi.nlm.nih.gov/pubmed/30858428
2018 July
Area perimeter intensity ImageJ script
The script is run in ImageJ (New>Script>Open>”path of script”). User selects the location containing no other files than acquired image files. The script works on Zeiss (.czi) files, but might work on other image file types. The images are automaticaly opened and the user is asked for the number of cells to be selected. Each selected cell is individually duplicated. On each duplicate the parameters of phalloidin channel such as intensity value, area etc are measured based on GLOBAL thresholding within duplication area. On each duplicate the parameters of pTyr channel such as intensity value, area etc are measured based on LOCAL thresholding within duplication area. The script runs all the images in the folder. The script creates a folder “processed” which contains the composite image with marked global and local thresholding of each selected cell in the image. The script produces a “results.txt” file, that contains all the measured parameters, each row representing one selection with variables separated by “;”. The “results.txt” file can be opened and analysed further in programs such as MS Excell. The user can compare the selections marked in composite image with the values in the “results.txt” as they are marked with unique matching identifiers based on original image file names.
Script author: Vid Šuštar
Project link at GitHub: https://github.com/mattilalab/MiMB/tree/master/Sustar_et_al_2018
Šuštar V, Vainio M, Mattila PK. Visualization and quantitative analysis of the actin cytoskeleton upon B cell activation. Methods in Molecular Biology. 2018;1707:243-257.
doi: 10.1007/978-1-4939-7474-0_18
PubMed link: https://www.ncbi.nlm.nih.gov/pubmed/29388113